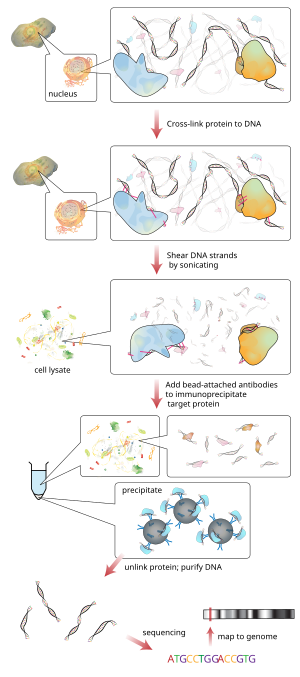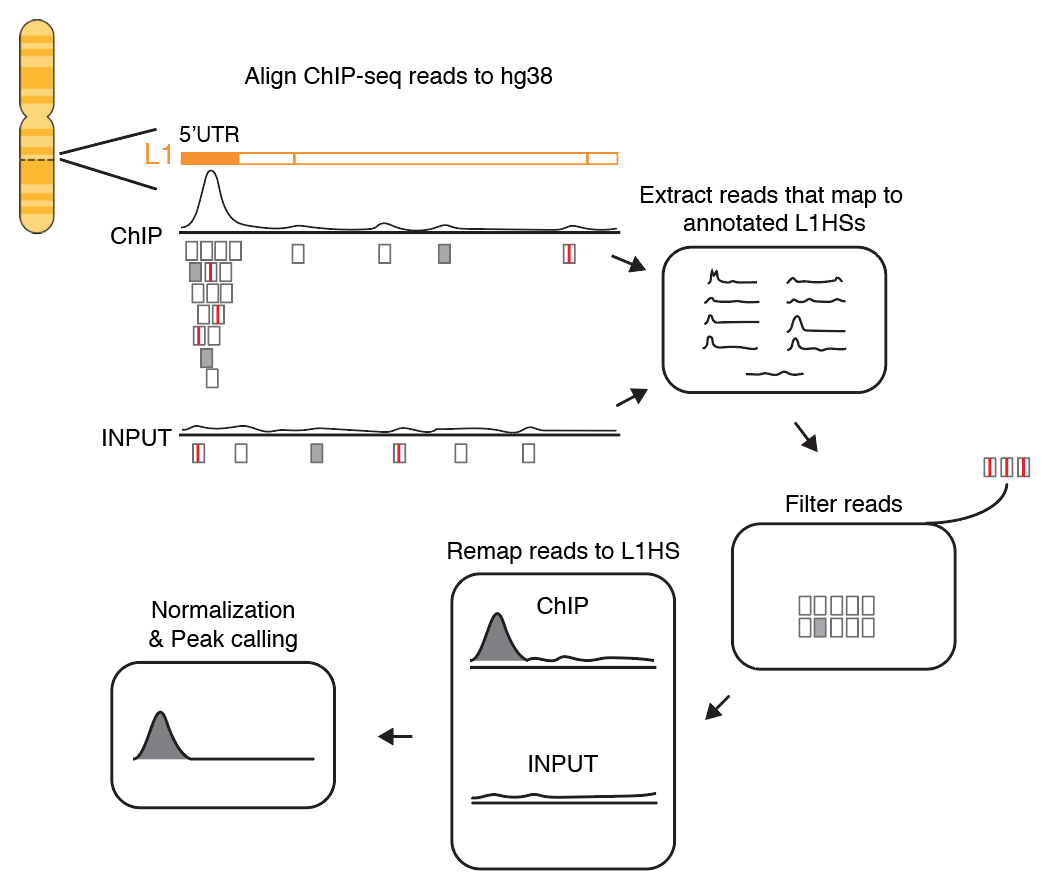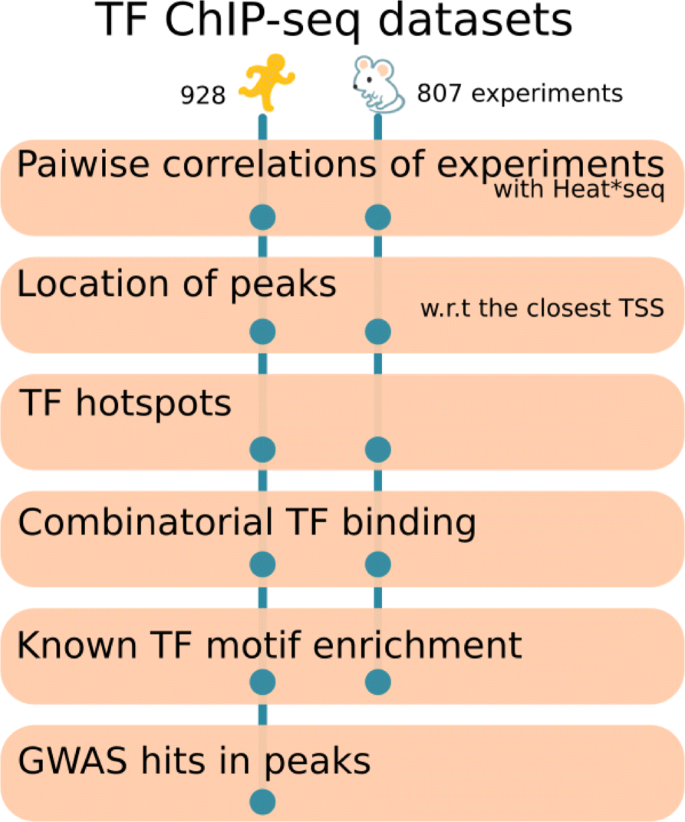
Insights into mammalian transcription control by systematic analysis of ChIP sequencing data | BMC Bioinformatics | Full Text
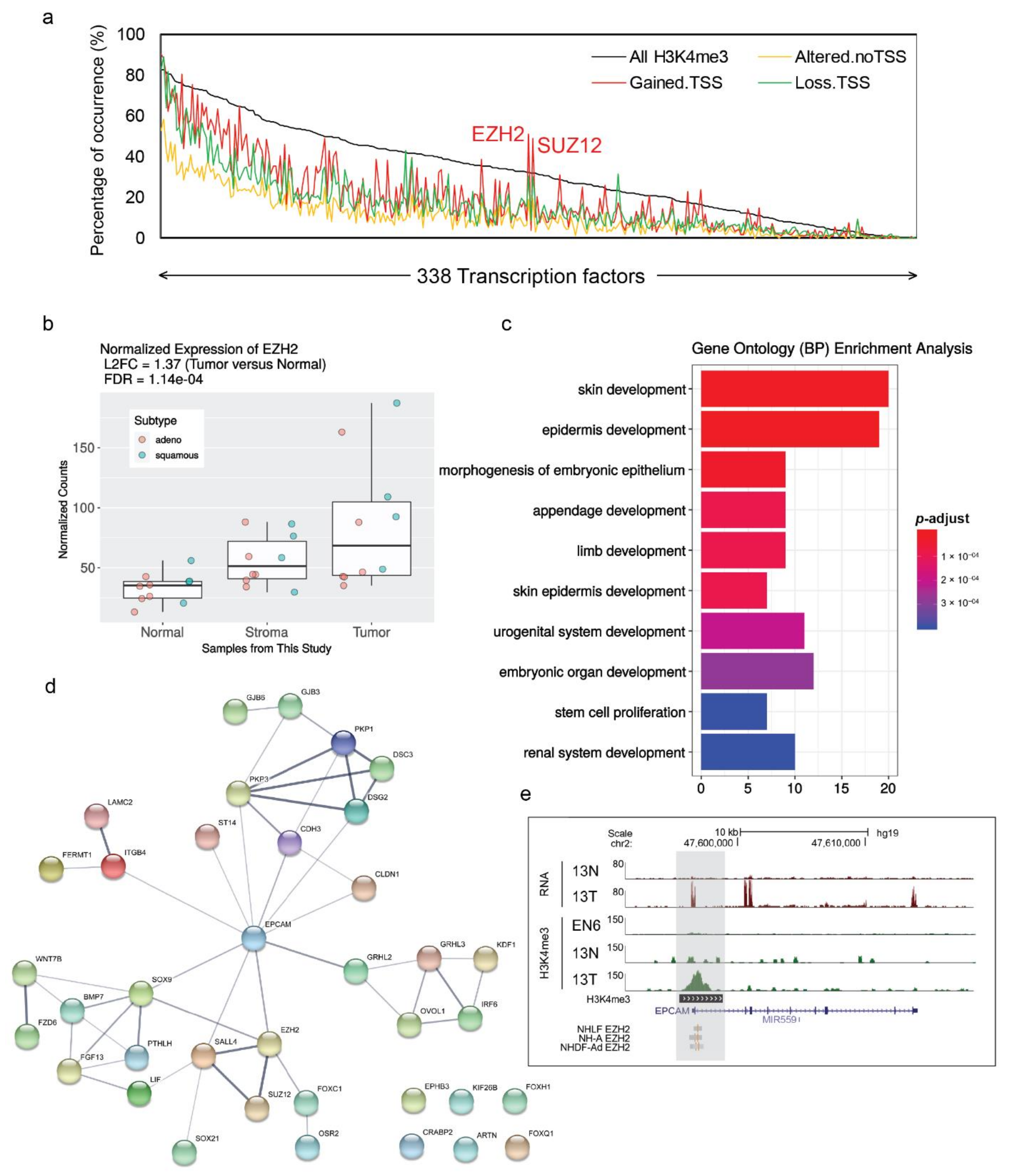
Cancers | Free Full-Text | Integrative RNA-Seq and H3 Trimethylation ChIP- Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection
![PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/68bd3de081d1baa8df24bec6ec2ad46df5e48b6c/5-Figure2-1.png)
PDF] Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse | Semantic Scholar

ChIP-Seq profiles obtained for ACF1 and RSF-1 in wild-type and mutant... | Download Scientific Diagram
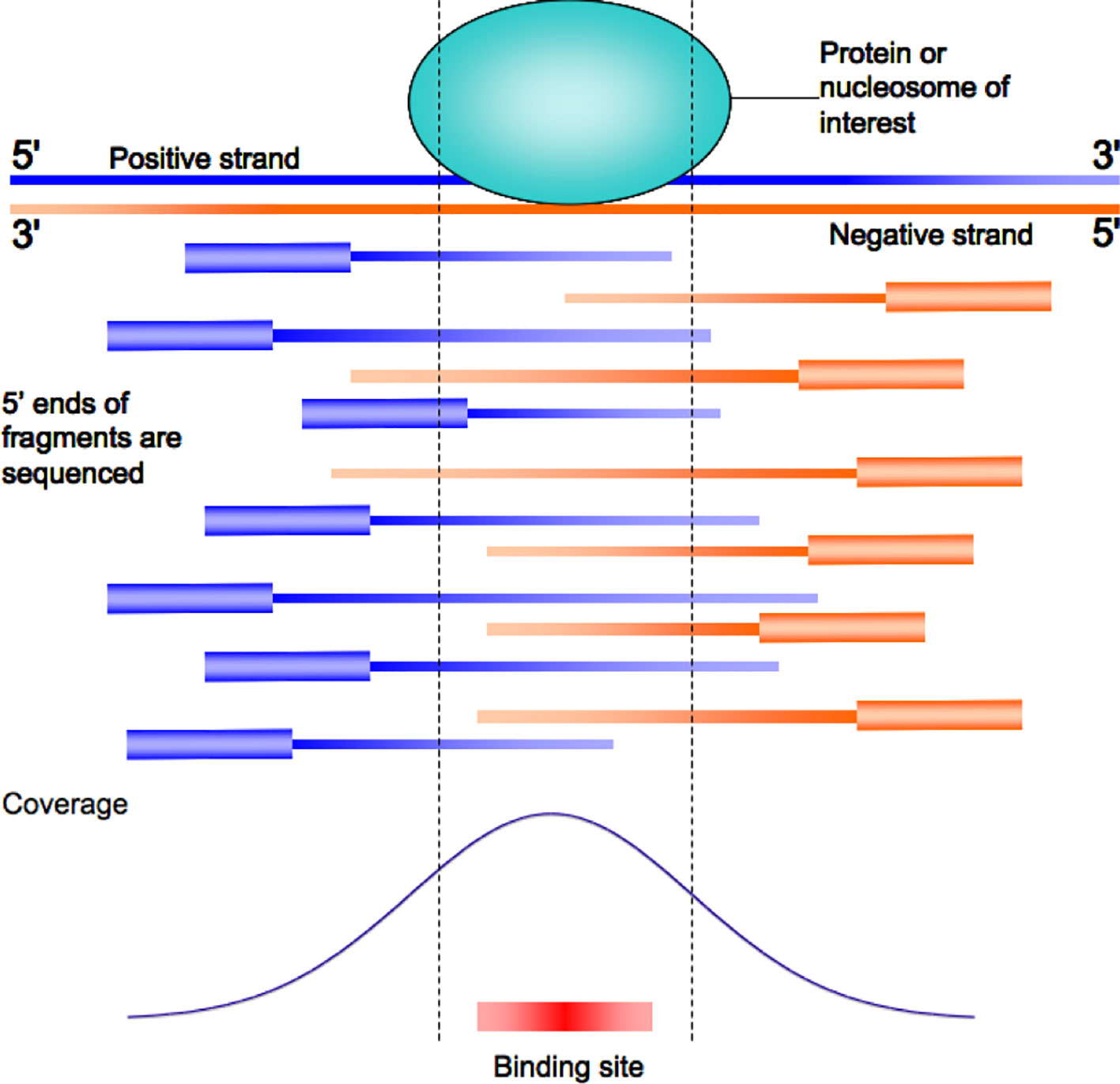
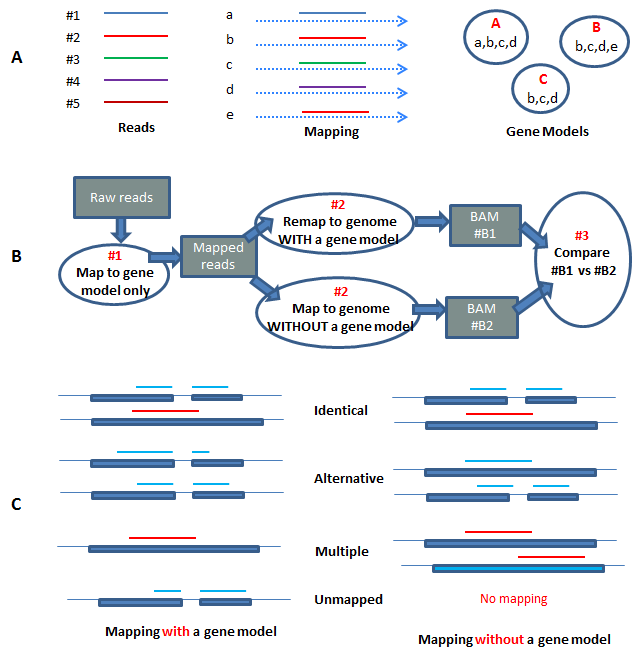
Frontiers | Integrating Peak Colocalization and Motif Enrichment Analysis for the Discovery of Genome-Wide Regulatory Modules and Transcription Factor Recruitment Rules

A comprehensive resource for retrieving, visualizing, and integrating functional genomics data | Life Science Alliance

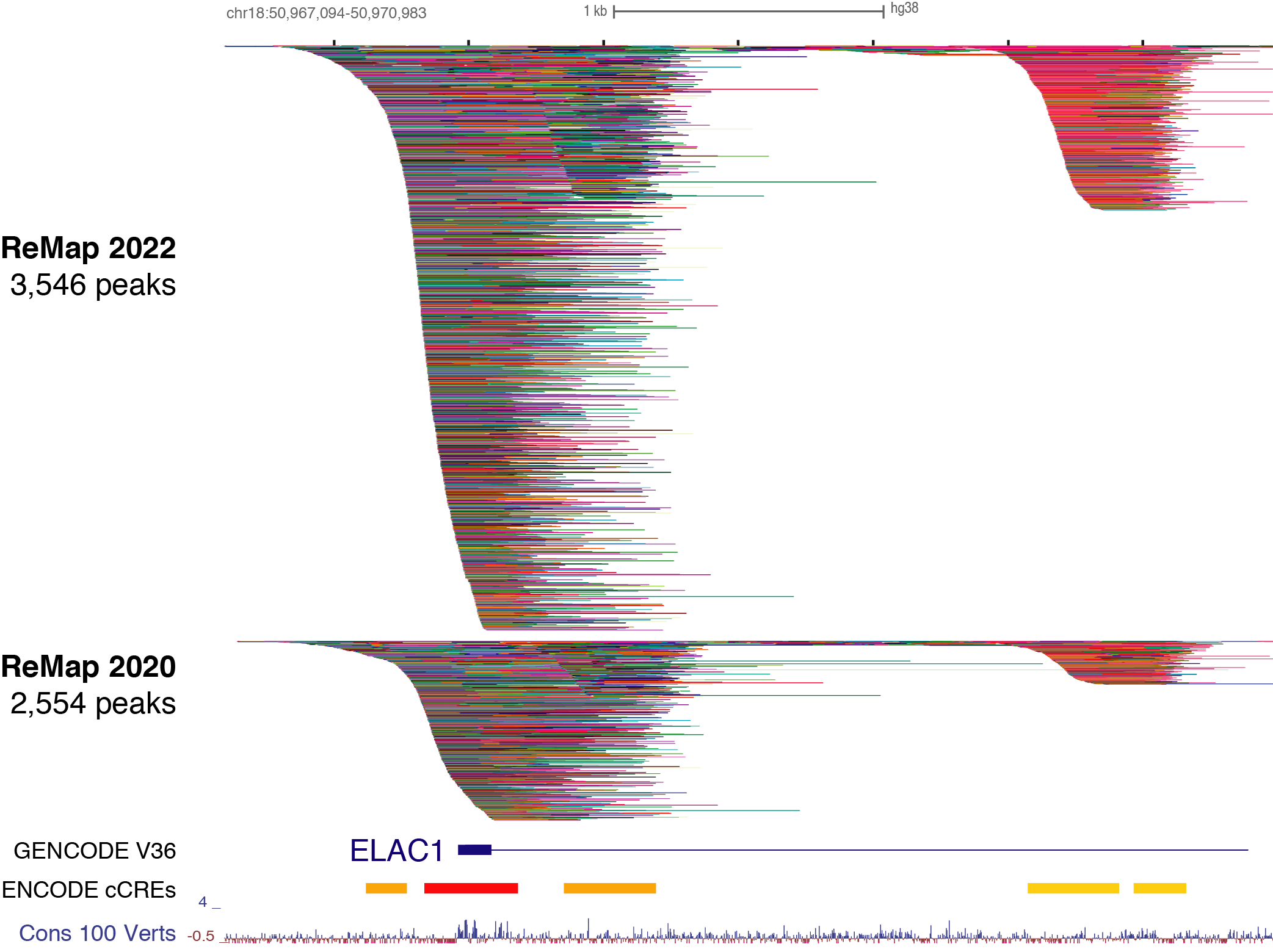
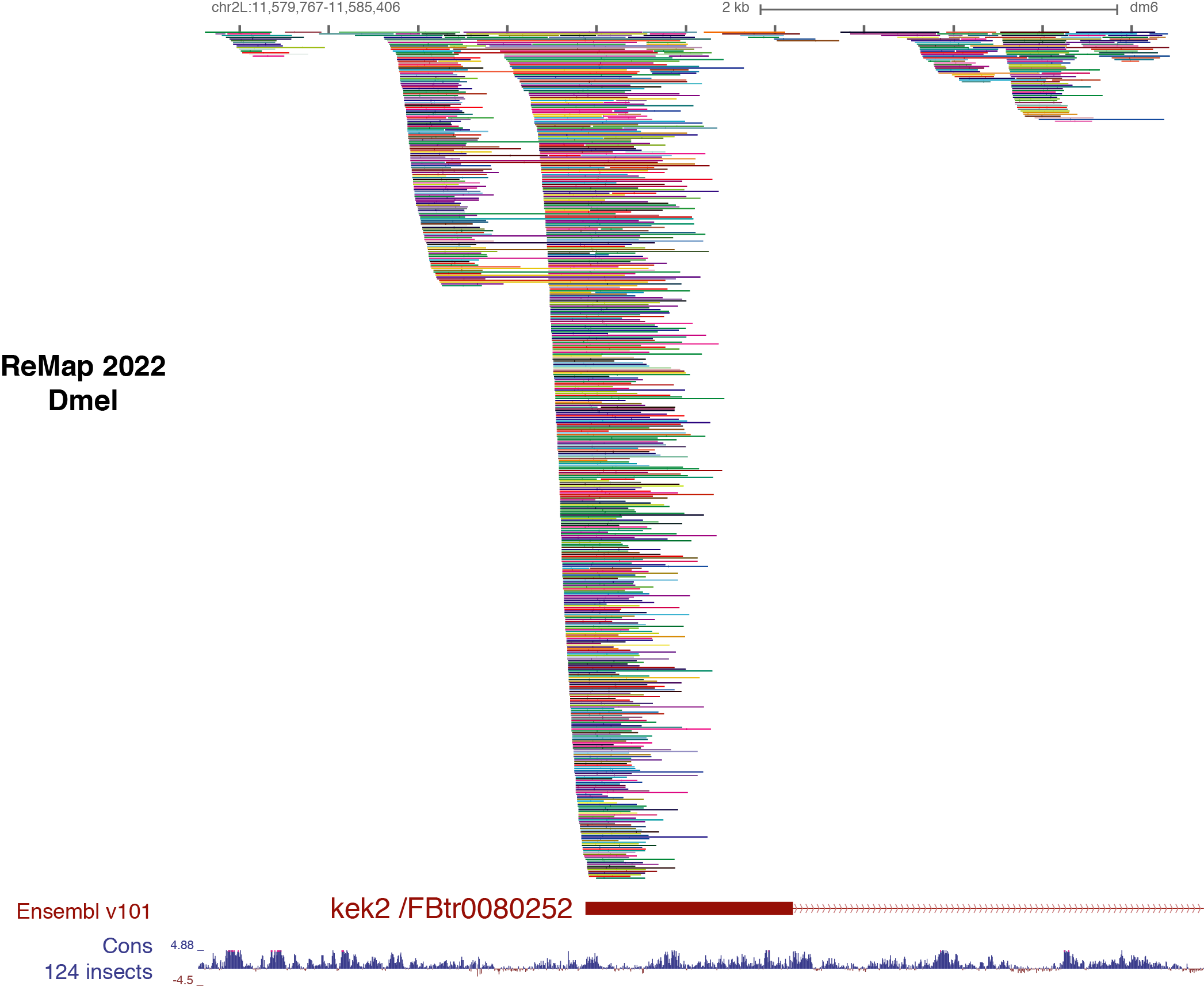
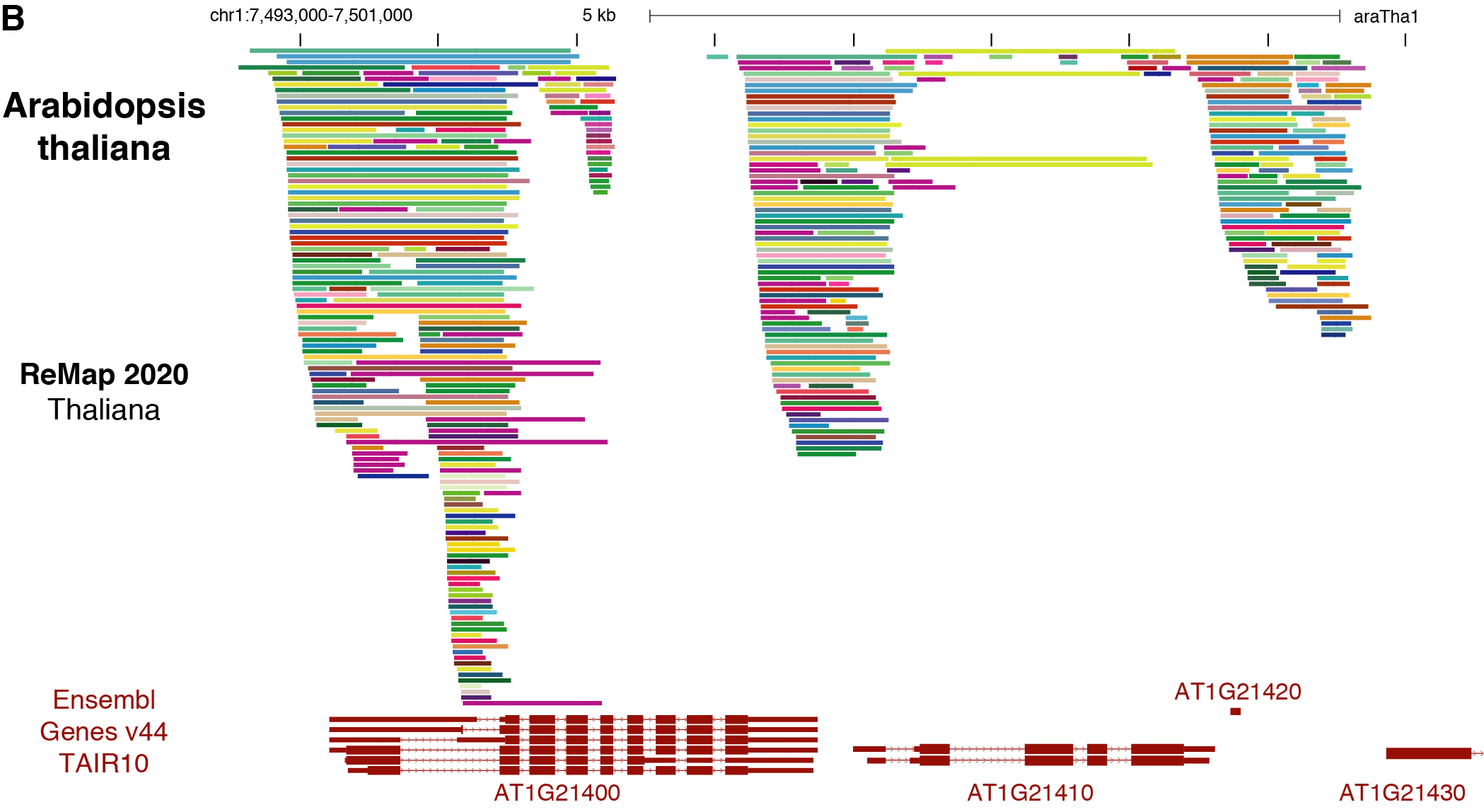
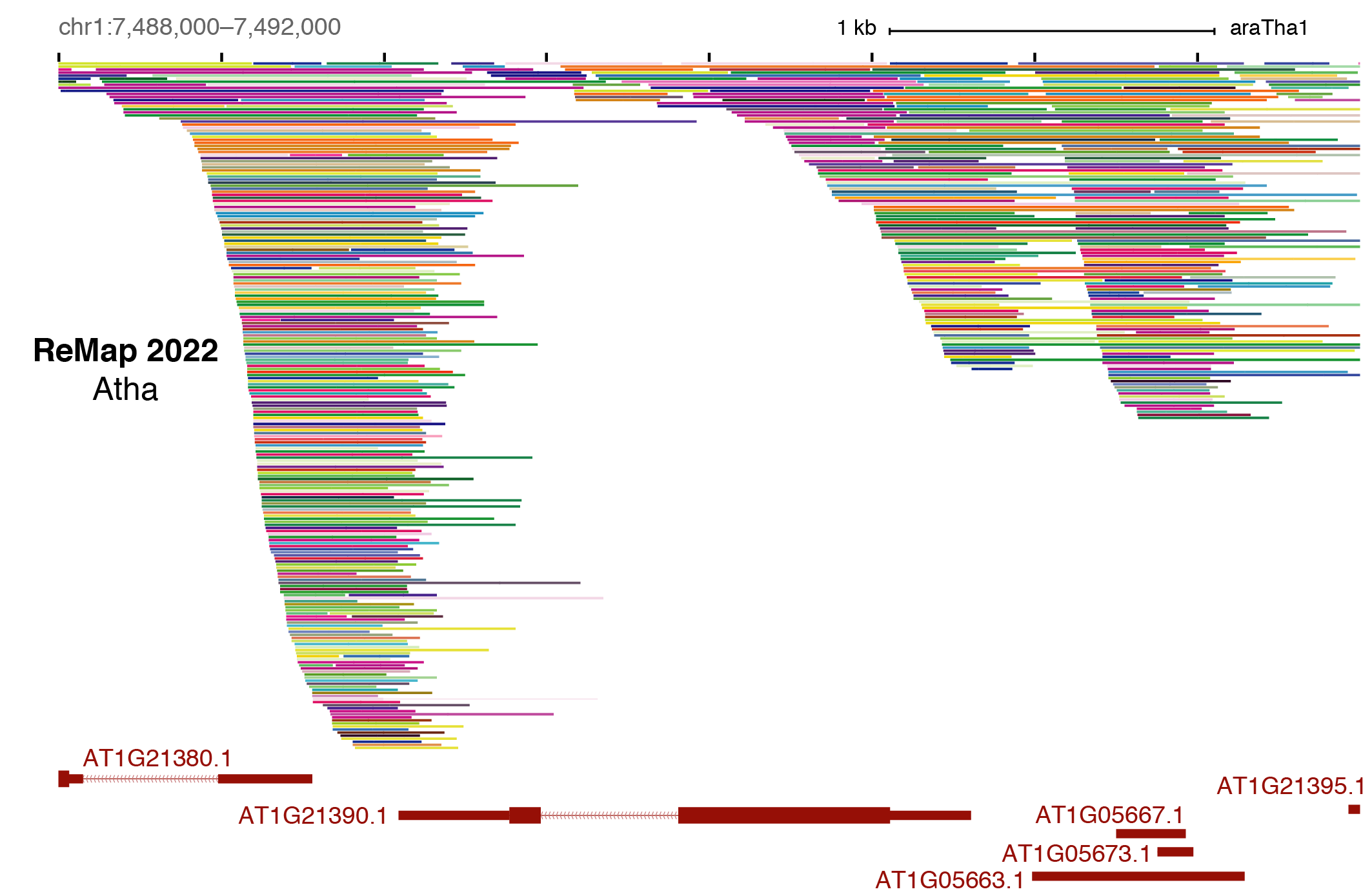
Benoit Ballester on Twitter: "The ReMap 2022 release is now on-line at https://t.co/3bv9RdYRfn. Regulatory ChIP-seq catalogue have been updated for Human and Arabidopsis, with new Mouse and Drosophila regulatory catalogues. https://t.co/m6jwUPyucS" /

Interrogating cell type-specific cooperation of transcriptional regulators in 3D chromatin - ScienceDirect