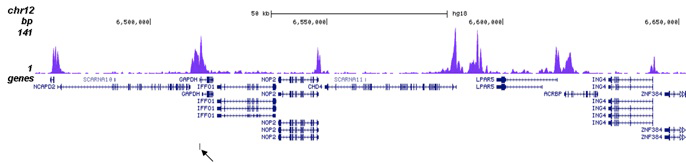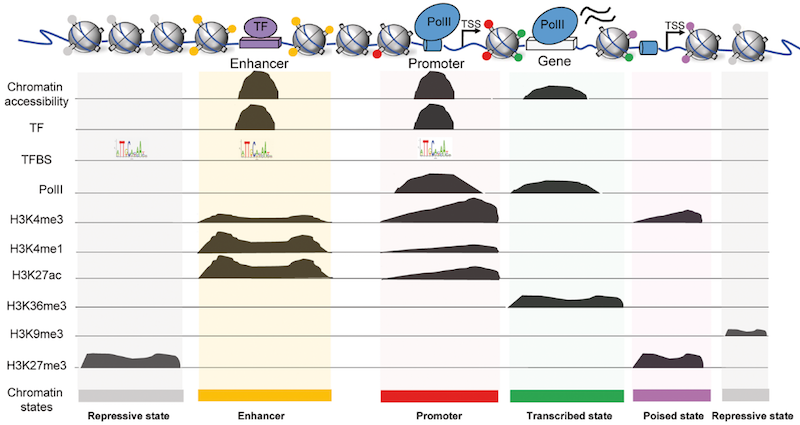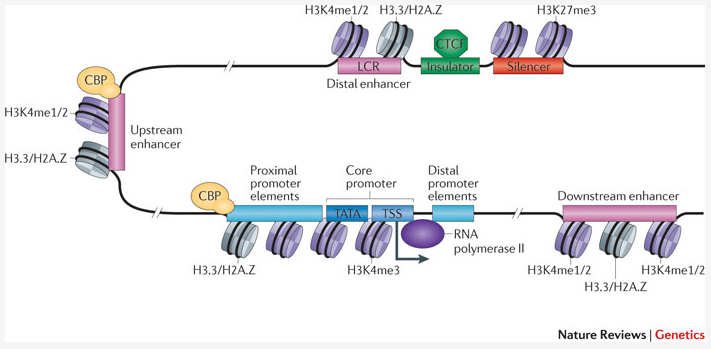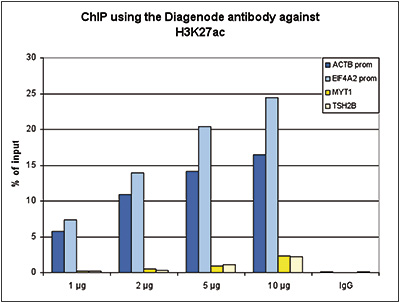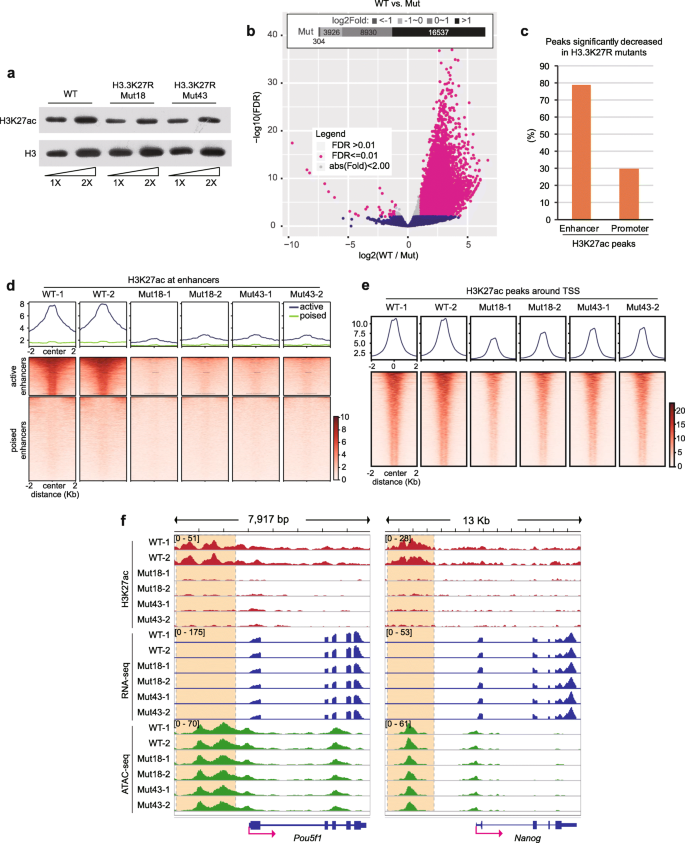
Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells | Genome Biology | Full Text

Capture‐C reveals preformed chromatin interactions between HIF‐binding sites and distant promoters | EMBO reports
Discovery of genes required for body axis and limb formation by global identification of retinoic acid–regulated epigenetic marks | PLOS Biology
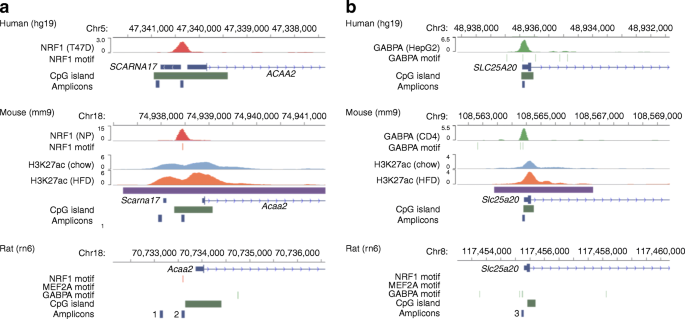
Genome-wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet-induced obesity in mice | SpringerLink

Histone H3K4me1 and H3K27ac play roles in nucleosome depletion and eRNA transcription, respectively, at enhancers | bioRxiv
Genome-Wide Analysis of Histone Modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica
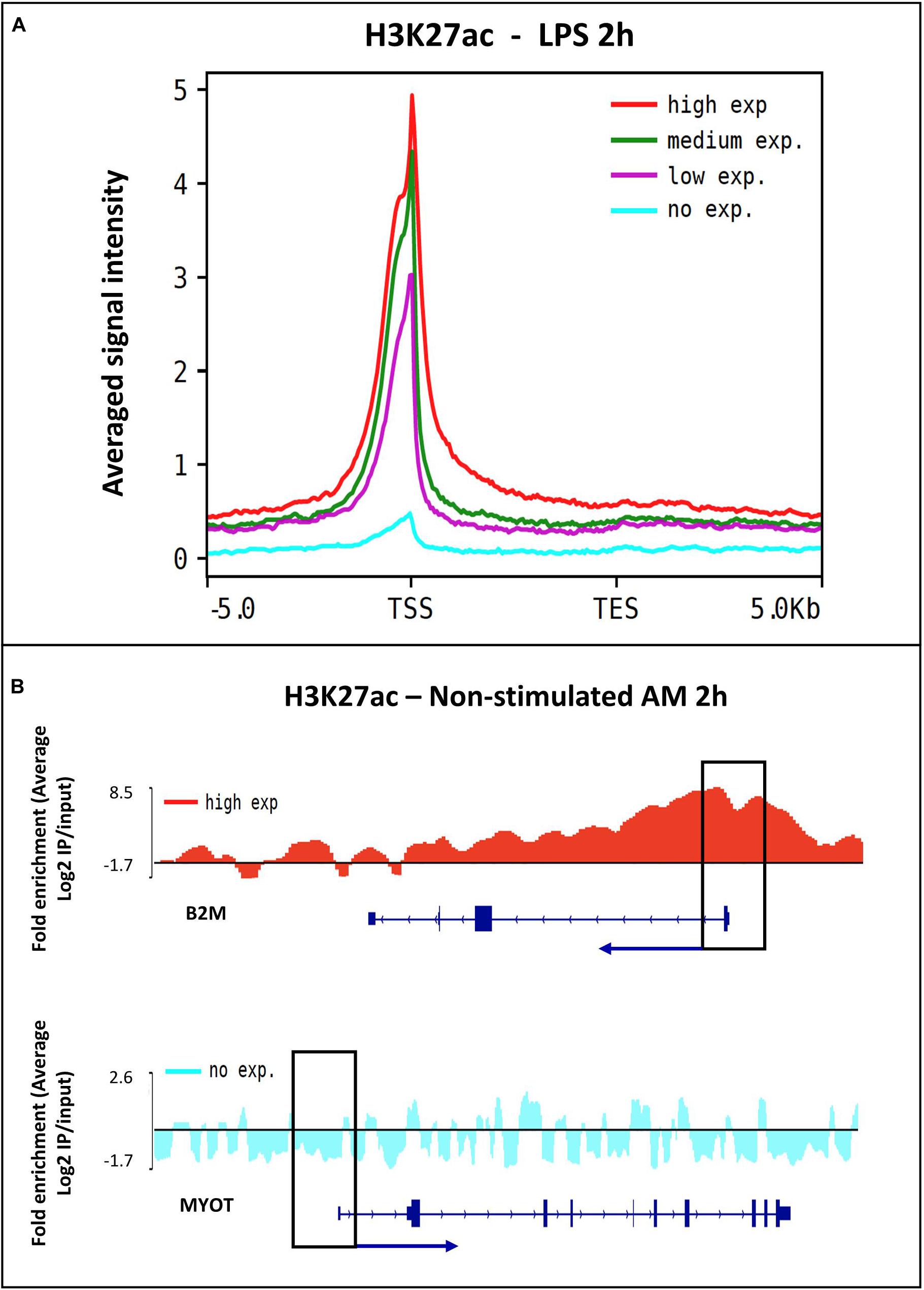
Frontiers | Changes in H3K27ac at Gene Regulatory Regions in Porcine Alveolar Macrophages Following LPS or PolyIC Exposure

Integrated Analysis of Whole-Genome ChIP-Seq and RNA-Seq Data of Primary Head and Neck Tumor Samples Associates HPV Integration Sites with Open Chromatin Marks. - Abstract - Europe PMC
Very long intergenic non-coding RNA transcripts and expression profiles are associated to specific childhood acute lymphoblastic leukemia subtypes | PLOS ONE
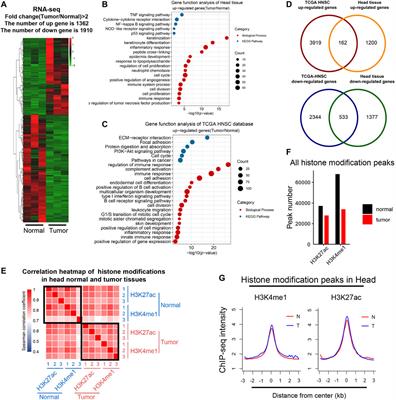
Frontiers | Genome-Wide Enhancer Analysis Reveals the Role of AP-1 Transcription Factor in Head and Neck Squamous Cell Carcinoma
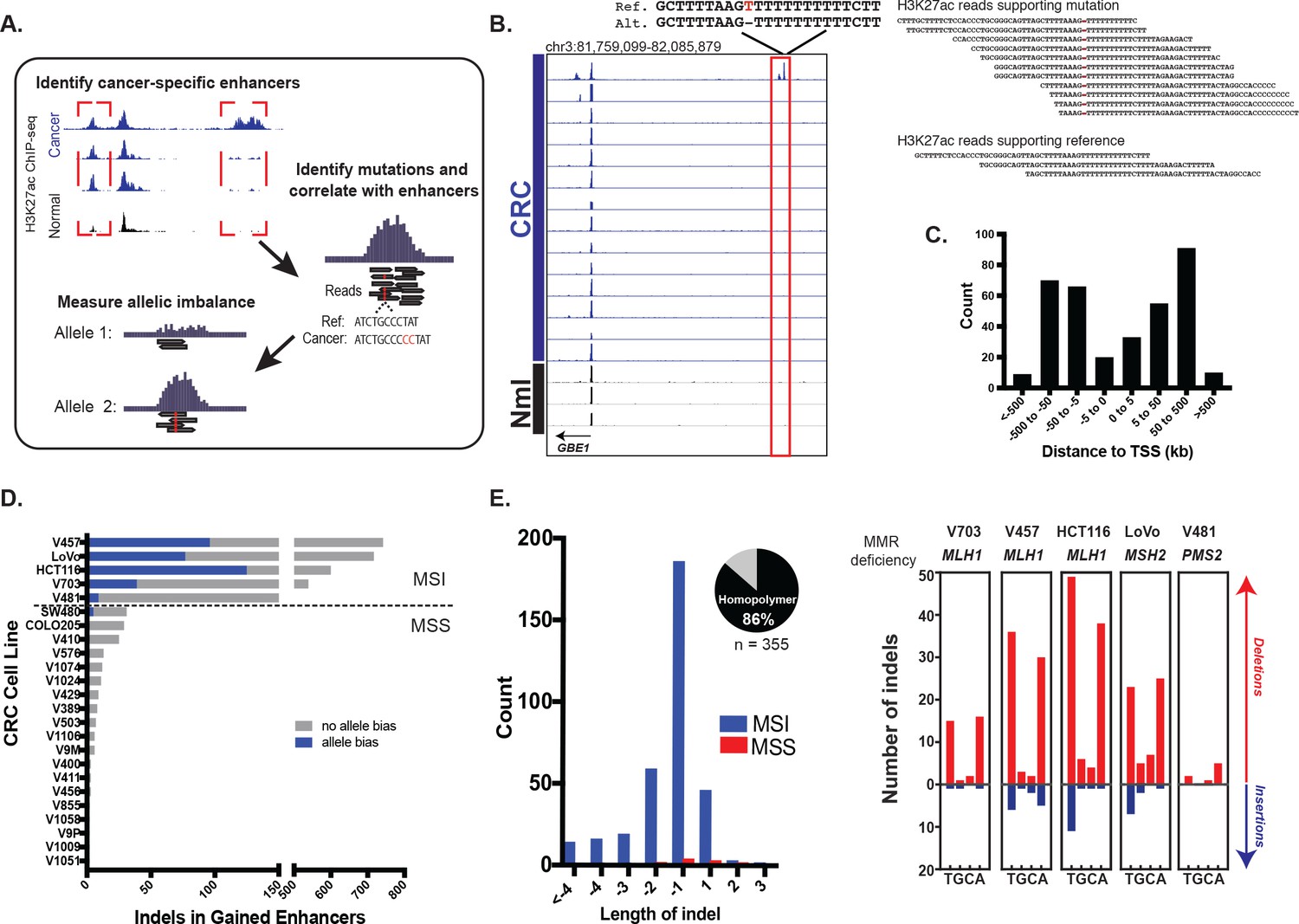
Figures and data in Mismatch repair-signature mutations activate gene enhancers across human colorectal cancer epigenomes | eLife

Poised and active enhancers. (A) H3K4me1 and H3K27Ac ChIP-seq tags from... | Download Scientific Diagram

FiTAc-seq: fixed-tissue ChIP-seq for H3K27ac profiling and super-enhancer analysis of FFPE tissues | Nature Protocols